CV – Research – Lab members – Publications
 Dr. Harold MacGillavry
Dr. Harold MacGillavry
Cell Biology, Faculty of Science, Utrecht University
Kruytgebouw, room N504
Padualaan 8, 3584 CH Utrecht
The Netherlands
Tel. +31-(0)30-2532722
e-mail: h.d.macgillavry@uu.nl
Curriculum Vitae
Harold MacGillavry studied Medical Biology at the University of Amsterdam, and obtained his PhD (2010) at VU University Amsterdam with Dr. Ronald van Kesteren, Prof. Dr. Guus Smit, and Prof. Dr. Joost Verhaagen. He then joined the lab of Dr. Tom Blanpied at the University of Maryland Baltimore (2010 – 2014). Here, he used live-cell, single-molecule imaging approaches to study the dynamic organization of neurotransmitter receptors and structural proteins at neuronal excitatory synapses. He demonstrated for the first time that the organization of proteins within individual synapses is highly non-uniform, with receptors and receptor-interacting proteins accumulating in distinct subsynaptic nanodomains. In 2013, he received an ALW-VENI grant from NWO and a Return-to-Europe fellowship from FEBS. He then joined the Cell Biology department at Utrecht University in 2014. Here, he investigates molecular mechanisms that control synaptic transmission. In 2016 he received a NARSAD Young Investigator Award, and a Starting Grant from the European Research Council (ERC).
Research Summary
We study the mechanisms that control the molecular organization and function of neuronal synapses. The goal of our lab is to reveal the dynamic molecular processes at synapses that control neuronal signaling. Insight in these processes is key for a fundamental understanding of brain function, but will also help understand the etiology of for instance autism spectrum disorders and neurodegenerative disorders such as Alzheimer’s disease.
The brain continuously integrates and processes sensory inputs through complex networks of neurons that are connected via specialized junctions, called synapses. At synaptic sites, electric impulses trigger the release of neurotransmitter from the presynaptic terminal that activate the neurotransmitter receptors retained in the postsynaptic membrane. Synaptic transmission allows neurons to communicate, and permits neurons to integrate into the neuronal circuits that encode and store information. Importantly, the strength of synaptic transmission can be adjusted in response to synaptic activity patterns. These plastic changes in synaptic strength are mediated by the trafficking of receptors to and from synaptic sites, events that underlie the long-term depression and potentiation of synaptic responses that are thought to be the cellular mechanisms of learning and memory.
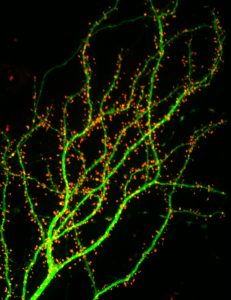
At excitatory synapses, multi-domain scaffolding proteins are organized in a dense macromolecular complex that defines the postsynaptic density (PSD), the major structural component of the synapse. These complexes are attached to the membrane where they can reversibly interact with receptors at the surface and effectively concentrate receptors close to the presynaptic release site. Our main interest is to investigate how scaffolding proteins and other synaptic components are organized to contribute to the retention of synaptic receptors at synaptic sites. We found that the distribution of synaptic proteins within individual synapses is highly heterogeneous, with scaffolds and receptors accumulating in distinct sub-synaptic nanodomains. This particular organization is likely to have important implications for synaptic physiology, and we have set out to determine the functional impact of this organization on synaptic signaling.
Currently, we investigate:
- the mechanisms that control the trafficking, motion and sub-synaptic distribution of synaptic proteins, and
- the functional contribution of these mechanisms to synaptic structure and signaling.
Approach
To study the molecular architecture and function of neuronal synapses at high temporal and spatial resolution, we use advanced live-cell confocal and super-resolution imaging techniques. In particular, we exploit single-molecule localization and tracking PALM (photoactivated localization microscopy) that allows us to investigate protein organization and motion within individual, living synapses at nanometer resolution. We are currently also exploring the possibilities that super-resolution STED (stimulated emission depletion) microscopy and FCS (fluorescence correlation spectroscopy) can offer. These advanced imaging approaches are combined with specific pharmacological and molecular interventions to gain mechanistic insight in the processes that control the molecular organization of synapses.
Lab Members
Postdoc
Yolanda Gutierrez – y.gutierrezgonzalo@uu.nl
Niels Reinders – n.r.reinders@uu.nl
PhD students
Wouter Droogers – w.j.droogers@students.uu.nl
Zehra Kazmi – s.z.h.kazmi@uu.nl
Nicky Scheefhals – n.scheefhals@uu.nl
Dimitrios Samouil – d.samouil@uu.nl
Publications
2017
Post H, Penning R, Fitzpatrick MA, Garrigues LB, Wu W, MacGillavry HD, Hoogenraad CC, Heck AJ, and Altelaar AF. Robust, Sensitive, and Automated Phosphopeptide Enrichment Optimized for Low Sample Amounts Applied to Primary Hippocampal Neurons. J Proteome Res, 2017; 16 (2): 728-737.
2016
Tang, A.H., Chen, H., Li, T.P., Metzbower, S.R., MacGillavry, H.D., and Blanpied, T.A. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210-214.
MacGillavry, H.D., Kerr, J.M., Kassner, J., Frost, N.A., and Blanpied, T.A. Shank-cortactin interactions control actin dynamics to maintain flexibility of neuronal spines and synapses. Eur J Neurosci 43, 179-193.
2015
MacGillavry, H.D., and Hoogenraad, C.C. The internal architecture of dendritic spines revealed by super-resolution imaging: What did we learn so far? Exp Cell Res 335, 180-186.
2014
Lu, H.E., MacGillavry, H.D., Frost, N.A., and Blanpied, T.A. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J Neurosci 34, 7600-7610.
2013
Frost, N.A., MacGillavry, H.D., Lu, H.E., and Blanpied, T.A. Live-cell PALM of intracellular proteins in neurons. In Nanoscale Imaging of Synapses, pp. 93-123.
MacGillavry, H.D., and Blanpied, T.A. Single-molecule tracking photoactivated localization microscopy to map nano-scale structure and dynamics in living spines. In Current Protocols in Neuroscience (John Wiley & Sons, Inc.).
MacGillavry, H.D., Song, Y., Raghavachari, S., and Blanpied, T.A. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615-622.
2012
Verhaagen, J., Van Kesteren, R.E., Bossers, K.A., Macgillavry, H.D., Mason, M.R., and Smit, A.B. Molecular target discovery for neural repair in the functional genomics era. Handb Clin Neurol 109, 595-616.
2011
MacGillavry, H.D., Kerr, J.M., and Blanpied, T.A. Lateral organization of the postsynaptic density. Mol Cell Neurosci 48, 321-331.
MacGillavry, H.D., Cornelis, J., van der Kallen, L.R., Sassen, M.M., Verhaagen, J., Smit, A.B., and van Kesteren, R.E. Genome-wide gene expression and promoter binding analysis identifies NFIL3 as a repressor of C/EBP target genes in neuronal outgrowth. Mol Cell Neurosci 46, 460-468.
Geeven, G., MacGillavry, H.D., Eggers, R., Sassen, M.M., Verhaagen, J., Smit, A.B., de Gunst, M.C., and van Kesteren, R.E. LLM3D: a log-linear modeling-based method to predict functional gene regulatory interactions from genome-wide expression data. Nucleic Acids Res 39, 5313-5327.
van Kesteren, R.E., Mason, M.R., MacGillavry, H.D., Smit, A.B., and Verhaagen, J. A gene network perspective on axonal regeneration. Front Mol Neurosci 4, 46.
2009-2004
MacGillavry, H.D., Stam, F.J., Sassen, M.M., Kegel, L., Hendriks, W.T., Verhaagen, J., Smit, A.B., and van Kesteren, R.E. NFIL3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. J Neurosci 29, 15542-15550.
2004 – 2008
Court, F.A., Hendriks, W.T., MacGillavry, H.D., Alvarez, J., and van Minnen, J. (2008). Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci 28, 11024-11029.
Stam, F.J., MacGillavry, H.D., Armstrong, N.J., de Gunst, M.C., Zhang, Y., van Kesteren, R.E., Smit, A.B., and Verhaagen, J. (2007). Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur J Neurosci 25, 3629-3637.
Derksen, P.W., Tjin, E., Meijer, H.P., Klok, M.D., MacGillavry, H.D., van Oers, M.H., Lokhorst, H.M., Bloem, A.C., Clevers, H., Nusse, R., et al. (2004). Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc Natl Acad Sci U S A 101, 6122-6127.






 Anna Akhmanova:
Cellular Dynamics
Anna Akhmanova:
Cellular Dynamics Lukas Kapitein: Biophysics
Lukas Kapitein: Biophysics Florian Berger:
Theoretical Biophysics
Florian Berger:
Theoretical Biophysics Agathe Chaigne:
Cell division dynamics
Agathe Chaigne:
Cell division dynamics Ginny Farías:
Neuronal Organelle Dynamics
Ginny Farías:
Neuronal Organelle Dynamics Martin Harterink:
C. elegans neurobiology
Martin Harterink:
C. elegans neurobiology Casper Hoogenraad: Molecular Neuroscience
Casper Hoogenraad: Molecular Neuroscience Maarten Kole:
Axonal Signalling
Maarten Kole:
Axonal Signalling Harold MacGillavry:
Synapse organization
Harold MacGillavry:
Synapse organization Sabrina Oliveira:
Molecular Targeted Therapies
Sabrina Oliveira:
Molecular Targeted Therapies Ihor Smal: Image Analysis, Smart Microscopy and AI
Ihor Smal: Image Analysis, Smart Microscopy and AI Frederik Verweij:
Extracellular Vesicle Biology
Frederik Verweij:
Extracellular Vesicle Biology